December 27, 2024 by David Appell , Phys.org
Collected at: https://phys.org/news/2024-12-scientists-machine-molecular.html
In an era of medical care that is increasingly aiming at more targeted medication therapies, more individual therapies and more effective therapies, doctors and scientists want to be able to introduce molecules to the biological system to undertake specific actions.
Examples are gene therapy and drug delivery, which for widespread use need to be both effective and inexpensive. In service of this goal, a trio of researchers has used machine learning to design a way to remove molecules inside a molecular cage. Their study is published in Physical Review Letters.
The research, whose lead author is Ryan K. Krueger of Harvard University, but to which each co-author contributed equally, uses differentiable molecular dynamics to design complex reactions to direct the system to specific outcomes.
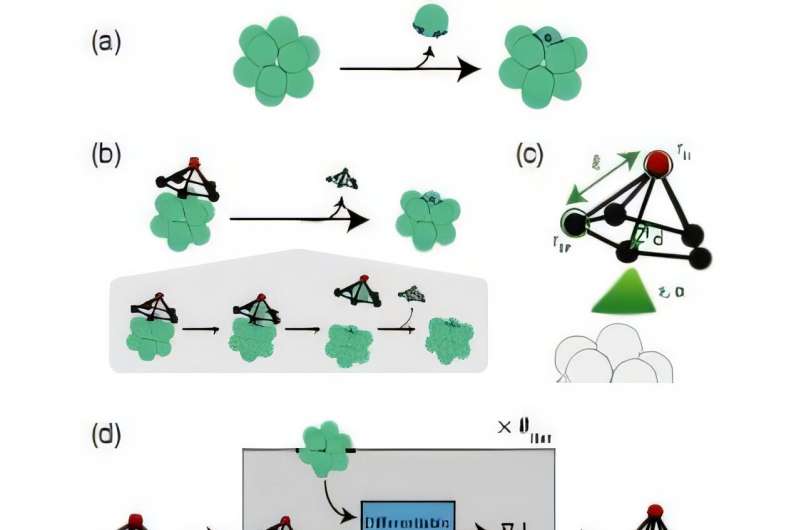
As an example, they undertook the controlled disassembly of colloidal structures—in particular, designing a molecule that could remove a particle surrounded and bound by a complete shell or “cage” of colloidal particles. (Colloids are mixtures of substances where nanoscopic or microscopic insoluble particles are dispersed throughout another substance. Examples are milk, smoke and gelatin.)
Machine learning was used to optimize the design of the shell’s “opener” molecule, which they call the “spider” due to its geometry. As they wrote, “disassembly is central to the dynamic functions of living systems, such as defect repair, self-replication, and catalysis.”
In particular, they designed for the controlled disassembly of icosahedral shells, collection of 12 particles with 30 outside edges connecting the shell particles. This configuration is much like protein capsids that house viruses.
The shell particles are considered “patchy”—their interactions with other shell particles, and the caged particle, have specific values of parameters that dictate the interaction’s directionality and relative strength. Introduced in soft material research 20 years ago, patchiness offers a versatile tunability in the designed interactions, achieving specific behaviors, assisted by the recent development of patchy particle simulations within a differentiable library.
Patchiness may even be varied over the surface of the patchy particles; here the 12 individual shell particles. The goal was to disassemble the shell, which carried an inherent tension between accomplishing the disassembly while maintaining the integrity of the substructure that remained.
The researchers assumed a Morse potential for the potential energy of the interacting shell particles, often used as a model of the interaction between the two atoms in a diatomic molecule, and with the caged molecule.
The Morse potential is simple and has three free parameters that can (and must) be selected for the desired situation. Removing the caged particle requires removing one of the shell particles.
For their analysis, the team assumed the object removing the shell particle was a rigid pyramid-type structure that would fit on top of the 12-sphere cluster. They called this object a “spider.” It consisted of a pentagon-shaped ring of particles that formed the base of the pyramid, with a single “head particle” on top of the pyramid assembly.
In their simulation, the icosahedral shell was given and fixed, with the spider free to land on any shell particle and interact with it.
The patch parameters were tuned so the spider as a whole was neither attracted or repelled by the cluster of shells, but the top-of-the-pyramid particle was attracted to patches on the shell particles by a force that could be varied by distance and strength. The dimensions of the spider and the radii of its head particle and base particles could also be adjusted.
Krueger and his collaborators used molecular dynamics, a standard technique which calculates the motion of each particle by the interaction forces it experiences with the other particles. They wanted to determine which particular parameters of the spider would pluck out the caged molecule from the shell.
Doing this on a computer by brute force—calculating for all possible parameters, particle by particle, until the desired outcome was reached—would take far too much computational power and time. So the group turned to machine learning to minimize a loss function that represented the tension between the disassembly and the remaining substructure integrity.
This process succeeded in producing a rigid spider that could accomplish the removal task. They then allowed the spider to flex, introducing a new free parameter that represented “configurable entropy.”
When it was optimized as well, the energy required to free the caged particle decreased. They found that a spider with asymmetrically flexible base legs required less energy to release the caged particle compared with a spider with the symmetrical, pentagonal base that was first assumed.
They noted their methodology can be broadly applied. “Since we optimize directly with respect to the numerically integrated dynamics, our method is general enough to study a wide range of systems,” they wrote.
“Foremost, it may enable experimental realizations of theoretical models that were otherwise limited by an inability to finely tune interaction energies.”
More information: Ryan K. Krueger et al, Tuning Colloidal Reactions, Physical Review Letters (2024). DOI: 10.1103/PhysRevLett.133.228201
Journal information: Physical Review Letters

Leave a Reply