Jordana Cepelewicz and John Rennie on October 4, 2017
This year’s Nobel Prize in Chemistry goes to researchers who made it possible to see proteins and other biomolecules at an atomic level of detail.
Seeing is believing, but for creatures as visually oriented as humans, seeing is also understanding. Much of the past century’s progress in biology has come from scientists being able to picture what DNA helices, protein channels and other biomolecules look like within the milieu of the cell. This year’s Nobel Prize in Chemistry celebrates three scientists for their role in developing cryo-electron microscopy, a technique that literally freezes biomolecules in living cells in mid-motion and resolves their images down to the level of atoms.
Jacques Dubochet at the University of Lausanne in Switzerland, Joachim Frank at Columbia University and Richard Henderson at the MRC Laboratory of Molecular Biology in Cambridge will share the Nobel Prize for work they pursued separately over decades to improve the state of biological imaging. Cryo-electron microscopy, usually called cryo-EM for short, is the brainchild of their contributions and those of their colleagues.
Proteins and other biomolecules are essentially nanomachines, and any detailed understanding of them requires knowing with precision how they change shape, bond to other molecules and pass around ions while sitting in cell membranes or floating in solution. The challenge of seeing them that way goes beyond overcoming their tininess. Chemical fixatives and stains can immobilize biomolecules for microscopy but they typically alter the form and configuration of the molecules in the process. Conventional freezing doesn’t help: Ice crystals rupture cell membranes and push the proteins around; the biomolecules react faster than the water freezes; and evaporating ice crystals disrupt the fidelity of the images.
In the mid-20th century, scientists began to use standard electron microscopy, X-ray crystallography and nuclear magnetic resonance imaging to study biomolecules, and those techniques overcame some of the problems, particularly with revealing the desired level of fine structure. But all of them traded off those answers for some level of accurate information about how the molecules looked and behaved in living systems.



In 1975, Henderson and his colleagues pushed electron microscopy to new heights in studies of bacteriorhodopsin, a purple bacterial photosynthetic pigment. The vacuum in an electron microscope normally wreaks havoc with cellular material by drying it out, but they stabilized the samples with a glucose solution. To avoid damaging the biological material, they illuminated samples with a much weaker electron beam than usual. Because the bacterial proteins cluster together tightly in the cell membrane, Henderson’s team could use X-ray crystallographic techniques to interpret the diffraction patterns of the weak electron beam.
As a result, they could resolve bacteriorhodopsin’s structure down to just 0.7 nanometers — the best yet for a protein. But that success was built on the peculiarly tolerant characteristics of the bacteriorhodopsin. A more generalizable approach was needed.
Frank provided part of that in the 1980s by devising a method for processing multiple 2D electron microscopy scans into high-resolution 3D images. Using the algorithms that he developed, computers could match the features of multiple protein molecules randomly positioned in a membrane and then infer a sharper structure for an average one of the protein molecules.
Meanwhile, Dubochet was improving on freezing as a way of immobilizing proteins. He realized that if the biological samples could be frozen quickly enough, the water in them would form a glass rather than crystalline ice, which would be less disruptive. He developed a technique that used liquid ethane and liquid nitrogen at -196 degrees C. to freeze samples for electron microscopy almost instantaneously. Dubochet’s work made cryo-EM a reality.
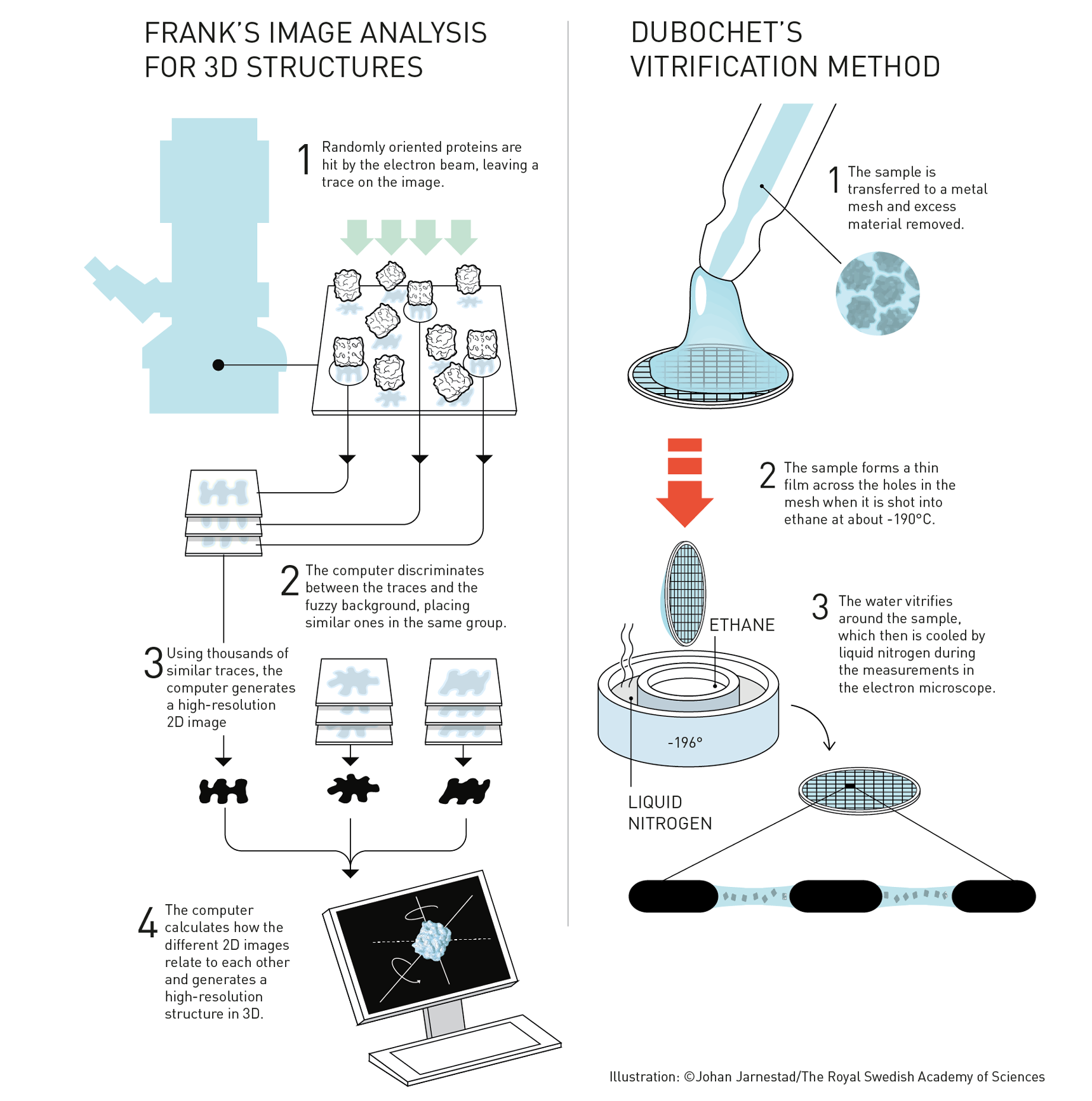
In 1991, Frank used Dubochet’s cryonic vitrification technique and his own software to produce 3D images of ribosomes with a 4-nanometer resolution. This remarkable but still blobby view of proteins was not enough, however, to satisfy Henderson, who was determined to see electron microscopy for proteins resolving individual atoms. He continued to push at improving the microscopic imaging technology. In 2013, he finally succeeded with a new, more sensitive detector that responds to single electrons directly.
Because of the transformative ease, simplicity and versatility of cryo-EM, it has become immensely useful to biological researchers in diverse fields. Essentially any laboratory in the world can now perform a type of molecular structural analysis that was once forbiddingly difficult if not impossible, according to Martin Beck, a senior scientist in the cell biology and biophysics unit of the Max Planck Institute of Biochemistry (EMBL Heidelberg). “And that has an enormous impact for people who want to do drug screenings, for those who want to understand how molecules function in disease, for those who want to understand HIV and cancer,” he said.
Collected at: https://www.quantamagazine.org/supercool-protein-imaging-gets-the-nobel-prize-20171004/
